SEGUID v2: Checksums for Linear, Circular, Single- and Double-Stranded Biological Sequences
This is work under construction! Please do not use this in production until we’ve officially released SEGUID v2.
The SEquence Globally Unique IDentifier (SEGUID) checksum (Babnigg and Giometti 2006) was introduced to provide a stable, unifying key for the same sequence in different databases facilitating linking protein sequences across databases. SEGUID v2 (Pereira et al. 2024) extends the original SEGUID method to support also double-stranded sequences (e.g. DNA) and circular sequences (e.g. proteins and double-stranded DNA).
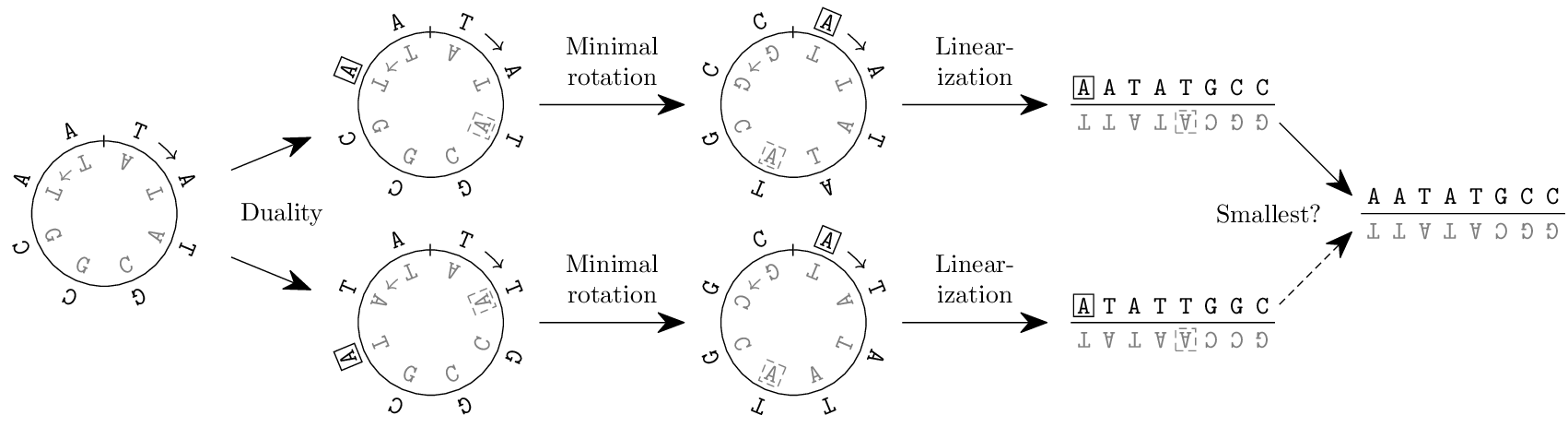
Example: SEGUID v2 for circular double-stranded DNA
SEGUID v2 is designed to be invariant to (i) rotation and (ii) duality (see above figure). No matter where we choose to “start” the circular dsDNA sequence, and no matter which strand we choose to be the Watson strand, the produced checksums are identical.
> const seguid = await import('./seguid.js')
> await seguid.cdseguid("TATGCCAA", "TTGGCATA")
'cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A'
## Same swapping Watson and Crick
> await seguid.cdseguid("TTGGCATA", "TATGCCAA")
'cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A'
## Same rotating two basepairs (= minimal rotation by Watson)
> await seguid.cdseguid("AATATGCC", "GGCATATT")
'cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A'>>> from seguid import *
>>> cdseguid("TATGCCAA", "TTGGCATA")
'cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A'
## Same swapping Watson and Crick
>>> cdseguid("TTGGCATA", "TATGCCAA")
'cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A'
## Same rotating two basepairs (= minimal rotation by Watson)
>>> cdseguid("AATATGCC", "GGCATATT")
'cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A'> library(seguid)
> cdseguid("TATGCCAA", "TTGGCATA")
[1] "cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A"
## Same swapping Watson and Crick
> cdseguid("TTGGCATA", "TATGCCAA")
[1] "cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A"
## Same rotating two basepairs (= minimal rotation by Watson)
> cdseguid("AATATGCC", "GGCATATT")
[1] "cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A"% source seguid
% puts [seguid::cdseguid "TATGCCAA" "TTGGCATA"]
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A
## Same swapping Watson and Crick
% puts [seguid::cdseguid "TTGGCATA" "TATGCCAA"]
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A
## Same rotating two basepairs (= minimal rotation by Watson)
% puts [seguid::cdseguid "AATATGCC" "GGCATATT"]
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A$ npx seguid --type=cdseguid <<< 'TATGCCAA;TTGGCATA'
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A
## Same swapping Watson and Crick
$ npx seguid --type=cdseguid <<< 'TTGGCATA;TATGCCAA'
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A
## Same rotating two basepairs (= minimal rotation by Watson)
$ npx seguid --type=cdseguid <<< 'AATATGCC;GGCATATT'
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A$ perl seguid --type=cdseguid <<< 'TATGCCAA;TTGGCATA'
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A
## Same swapping Watson and Crick
$ perl seguid --type=cdseguid <<< 'TTGGCATA;TATGCCAA'
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A
## Same rotating two basepairs (= minimal rotation by Watson)
$ perl seguid --type=cdseguid <<< 'AATATGCC;GGCATATT'
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A$ python -m seguid --type=cdseguid <<< 'TATGCCAA;TTGGCATA'
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A
## Same swapping Watson and Crick
$ python -m seguid --type=cdseguid <<< 'TTGGCATA;TATGCCAA'
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A
## Same rotating two basepairs (= minimal rotation by Watson)
$ python -m seguid --type=cdseguid <<< 'AATATGCC;GGCATATT'
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A$ Rscript -e seguid::seguid --type=cdseguid <<< 'TATGCCAA;TTGGCATA'
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A
## Same swapping Watson and Crick
$ Rscript -e seguid::seguid --type=cdseguid <<< 'TTGGCATA;TATGCCAA'
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A
## Same rotating two basepairs (= minimal rotation by Watson)
$ Rscript -e seguid::seguid --type=cdseguid <<< 'AATATGCC;GGCATATT'
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A$ tclsh seguid --type=cdseguid <<< 'TATGCCAA;TTGGCATA'
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A
## Same swapping Watson and Crick
$ tclsh seguid --type=cdseguid <<< 'TTGGCATA;TATGCCAA'
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44A
## Same rotating two basepairs (= minimal rotation by Watson)
$ tclsh seguid --type=cdseguid <<< 'AATATGCC;GGCATATT'
cdseguid=dUxN7YQyVInv3oDcvz8ByupL44AAvailability
Implementations of above SEGUID methods are currently available for JavaScript, Perl, Python, R, and Tcl;